For additional assistance, please contact support@metavilabs.com.
Imaging System Integration for Fully Automated Screening
Contents
Overview Security Installation Normal Usage
The FastTrack AI system can integrate with the Incucyte, Image Express, and many other systems to add full automation with advanced AI based analysis.
Overview
In custom projects, MetaVi Labs can install an ML-Compressor in the client's own lab. This is a machine that will copy images from the Incucyte Controller, compress them in a high quality compression format, and upload them into the FastTrack AI system for automated analysis.
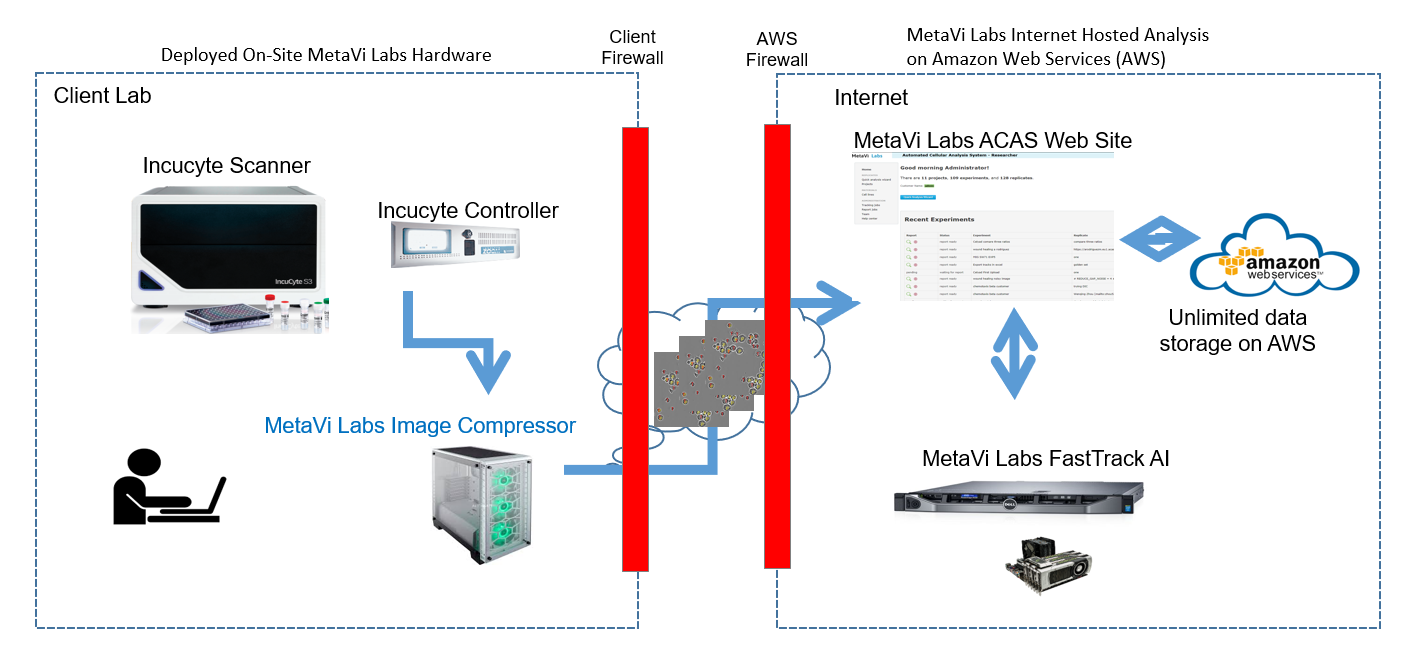
All data is stored on Amazon Web Services S3 cloud which provide security, integrity, and unlimited capacity.
MetaVi Labs license agreement only allows anonymous data on our service.
Experiment parameters such as time scales, resolution, and well conditions are stored in a data-base running on a linux VM on Amazon Web Services, protected by the AWS firewalls.
Security
Security is provided by several factors.
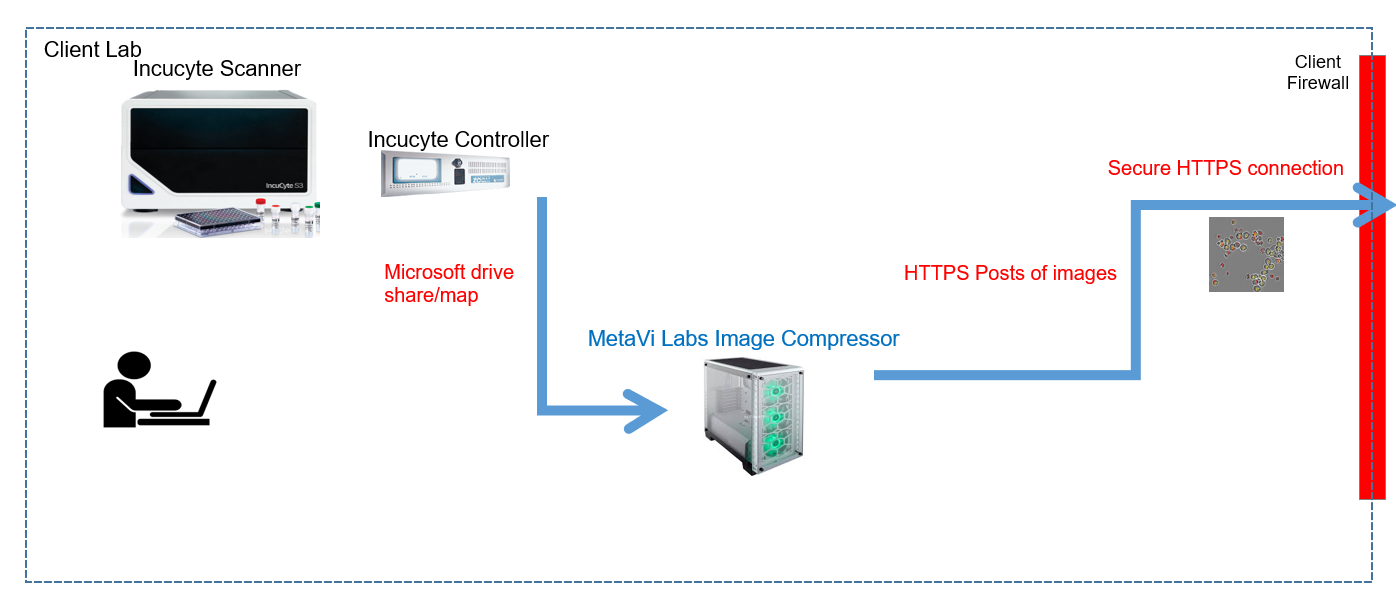
All Lab connections are initiated internally by client personnel. Uploads of images only occur when an authorized user initiates a capture session in the FastTrack AI User interface. And before any uploads can occur, the on-site compressor machine must have pre-installed a secure hash-code that is unique to the client's FastTrack AI account. At no time are inbound (into the client's intranet) connections attempted from our external web-services to the internal compressor unit. All connections are one-way, outbound (from the client's intranet). The compressor polls the web-service for capture jobs. This poll rate can be configured to a time period specified by the client.
All connections are encrypted via HTTPS. HTTPS is the internet standard for secure web connections (for example, used by banks for user webpage access to accounts). MetaVi Labs services provide X.509 certificates which have been signed by well-known certificate authorities (you will always see a lock in the URL when accessing FastTrack AI). The compressor software uses the same HTTPS interface to upload images as the user web page (transfers take place using a RESTful API).
All of our services are hosted on Amazon Web Services (AWS). Hosted servers and data are secured by Amazon hosting services at Amazon data centers. You can read more about AWS Security here: External link to AWS Security
Installation
The MetaVi Labs' compressor will arrive with pre-installed software. It will also be pre-configured for the client's account with the client's private account key (stored using the Microsoft .Net encryption library). MetaVi Labs will provide the client with the user-name and password.
Steps which must be taken by clients' IT staff:
- On the Incucyte, share with read-only permissions "\EssenFiles\ScanData" folder (this contains the raw images as recorded).
- On the ML Compressor, add the computer to the local domain so that it can find the Incucyte share.
- On the ML Compressor, open a windows explorer in the Administrator account and ensure that the path "\\zoom_system_name\EssenFiles\ScanData" is accessible.
- Edit the file (maybe D:\ or C:\) C:\MetaViLabs\config\harmony.config. Modify the capture-dir node to reflect the mount share drive of the Incucyte:
<capture-dir>\\zoom_system_name\EssenFiles\ScanData</capture-dir> - Edit the file C:\MetaViLabs\config\harmony.config. Modify the poll-timeout node to reflect the desired number of seconds between
poll events. This rate as which the Compressor will poll the web service looking for capture jobs that have been initiated by the user.
<poll-timeout>2</poll-timeout> - Run the program C:\MetaViLabs\Tools\SetupWizard.exe
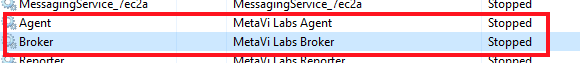
Click Start When prompted for the Access Key, enter the FastTrackAI account admin user's access key. When prompted for the AWS Instance ID, enter the computer MAC Address or IPV6 address (its a unique identifier). Click Exit - Open the Windows "Services" application and find the two MetaVi Labs services, and set both to Log On using "This Account: .\Administrator"
MetaVi Lab Agent MetaVi Lab Broker - Re-start the MetaVi Labs services (in Task Manager/Services, right-click, then start or restart):
Normal Usage
In day-to-day usage, the experimenter must log into their FastTrack AI account and initiate an experiment Capture. The on-site compressor periodically polls the FastTrack AI service to discover capture jobs that have been user initiated.
FastTrack AI organizes well recordings into Projects, Experiments, and Replicates.
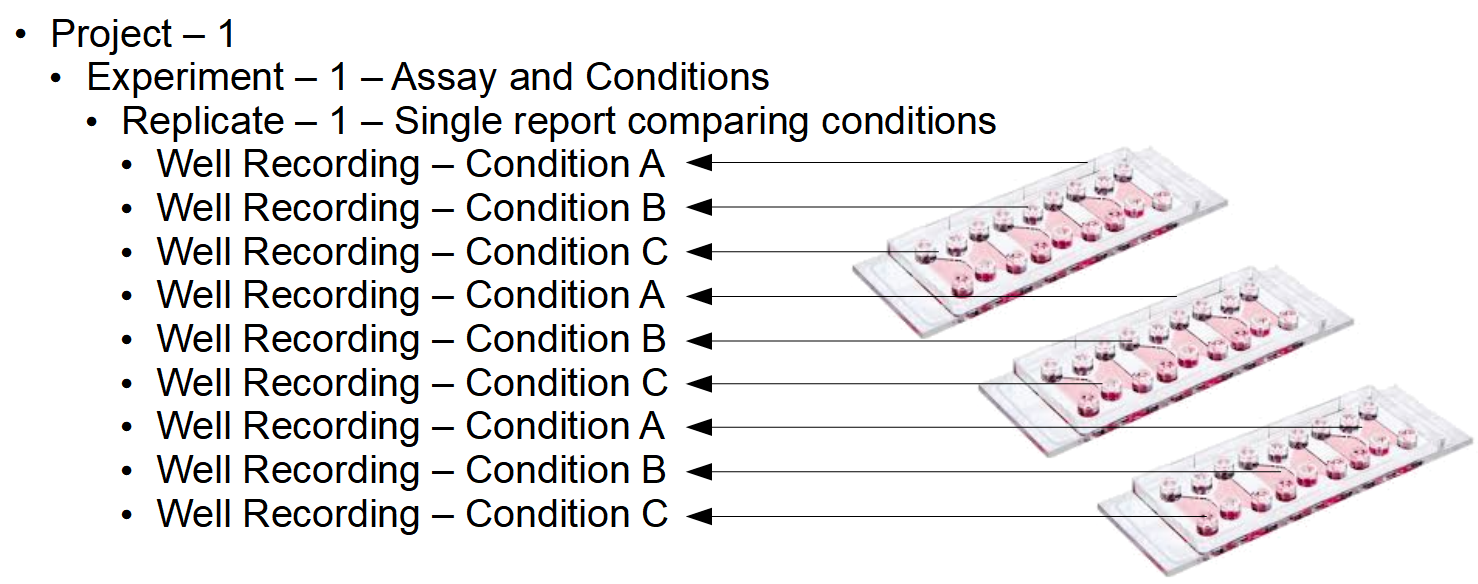
A project holds many experiments and the experimenter can choose to add other users in the organization to a project (read more about it in the Project page of the Help Center). An experiment holds parameters such as the plate map, time between frames, and other important information. An experiment also holds one or more replicates. A replicate is a repeat of the same experiment (no parameters have changed except the time and vessels). A report is computed for each replicate. All wells in a replicate are compared.
For the on-site Compressor to capture images, it must know the start time, end time, and vessel ID for the experiment. Once the experimenter clicks "start" in the FastTrack AI interface, the Compressor will watch the file system on the Incucyte Controller. When the last image is written, all images will be copied, compressed, and uploaded into the FastTrack AI system. Once the files are uploaded, analysis will automatically begin. Results will be available in a few hours.
The experimenter must setup an Incucyte experiment and configure a plate. The plate-map must be exported so that it can be uploaded into MetaVi Labs FastTrack AI. If the plate-map does not change for future experiments, the same exported file can be used for future replicates.
Normal Usage Step 1 - Creating a New Experiment
Open Projects from the left navigation bar, then either enter and existing project or create a new project. Inside the project, either enter an existing experiment, or create a new experiment (this same experiment can be re-used later). If entering an existing experiment, click the edit icon (pencil and paper icon). If creating a new experiment, the "+ Experiment" button will take you straight to the experiment edit page. In the experiment edit page, find the "+ plate map" button and upload the exported plate-map. Fill in the other values (specifics treated in specific assay guides). Then click save at the bottom.
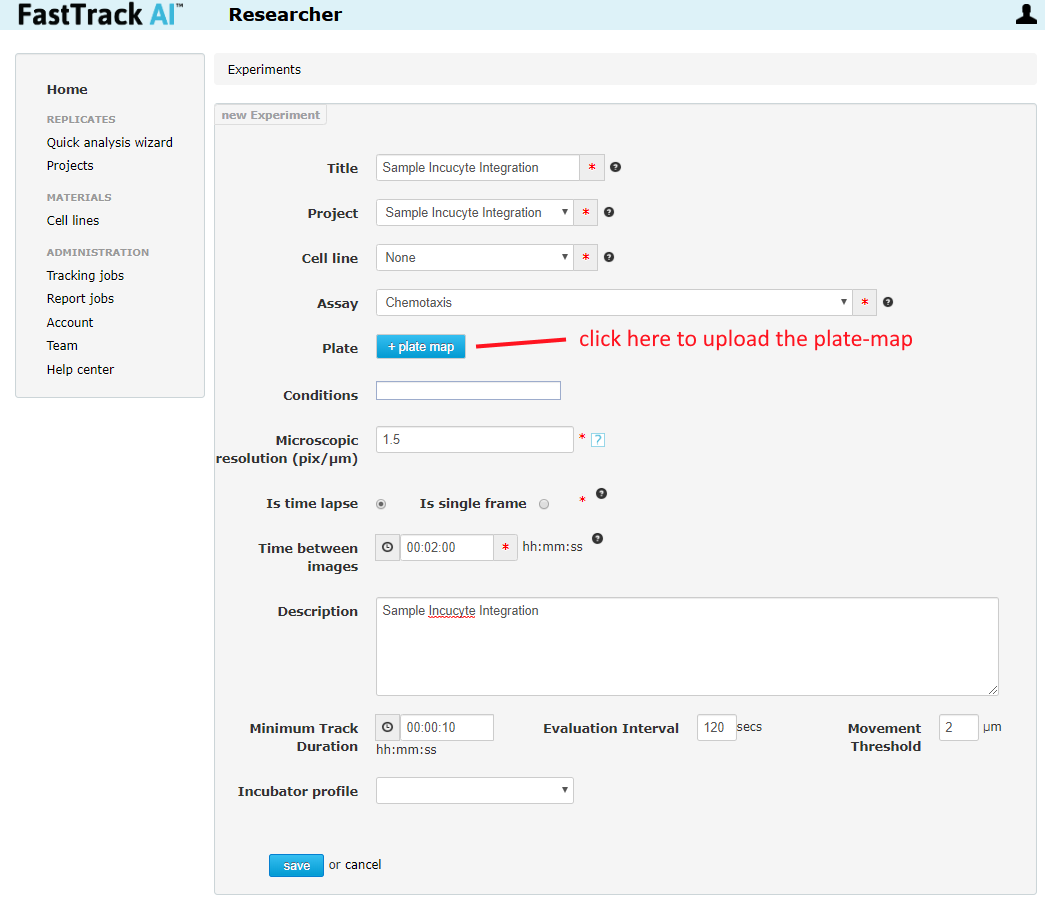
Normal Usage Step 2 - Creating a New Replicate
Once you save, the page will be directed to the newly created experiment. Inside the experiment page, click "+ replicate" to create a new replicate.
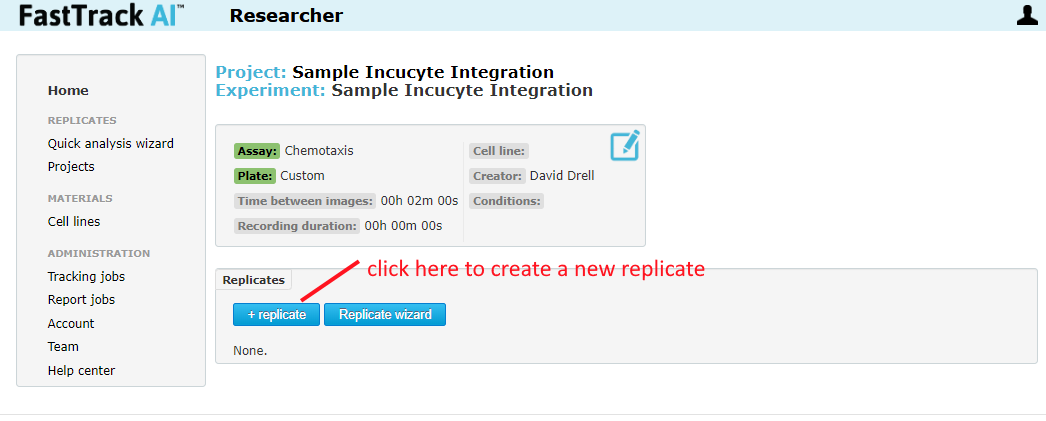
Normal Usage Step 3 - Entering Start/End Time
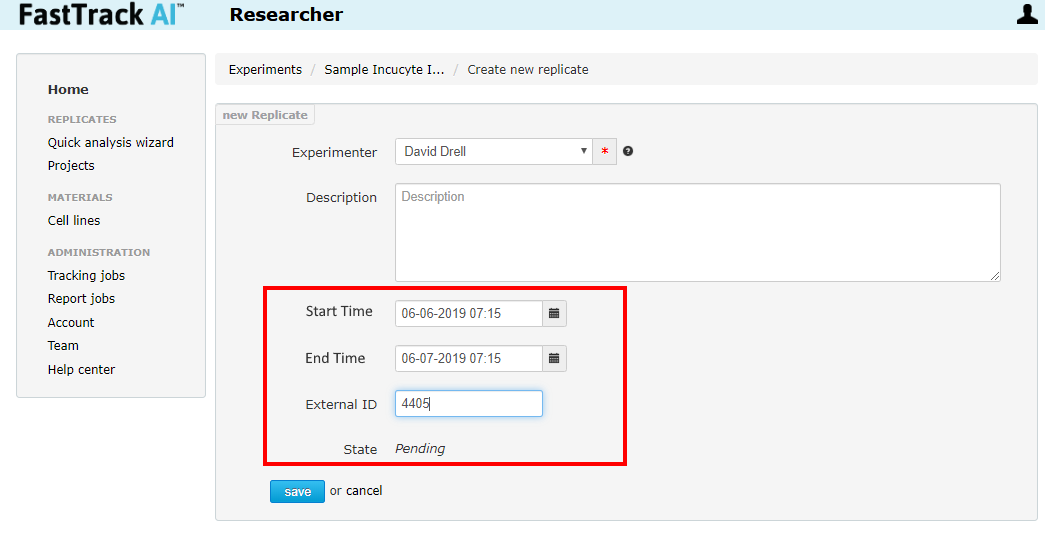
After clicking to create a new replicate, the page will be directed to the newly created replicate edit page. Find the start time, end time, and "External ID" fields. Enter the start and end time of the experiment. Uploading will not begin until the last image is recorded according to the stop time.
Note: the "External ID" field holds the vessel ID in the case of the Incucyte. In the case of ImageExpress, this field holds the custom "folder name" for the experiment run.
Click Save when done.
Normal Usage Step 4 - Start Analysis
After clicking to save a new replicate, the page will be directed to the newly created replicate. Find the "start" button. Click this to cause the Compressor to start monitoring for Incucyte images. Uploading will not begin until the compressor detects the final image has been created at the indicated stop time.
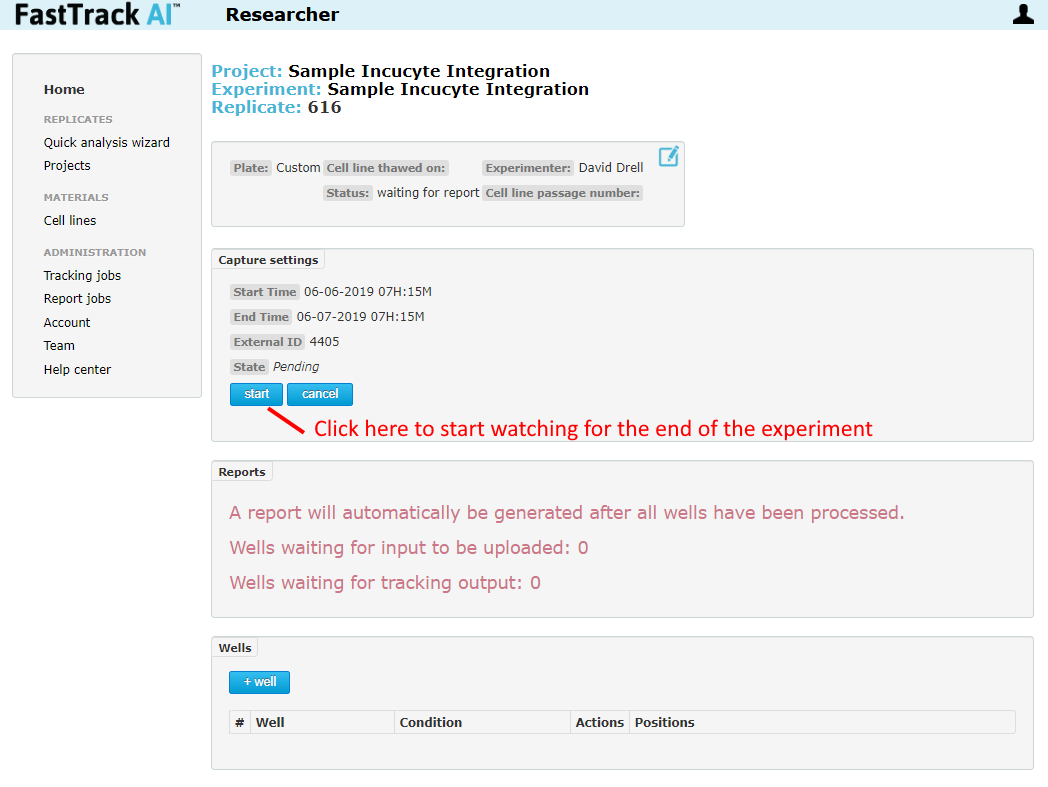
Normal Usage Step 5 - Getting Results
Thats it for the first experiment. The results will show up in a few minutes to a few hours depending on how many wells and how many frames per well were recorded. The report for each replicate will be inside the replicate.
Future experiments can be new replicates in the same parent Experiment (no parameters have changed), or you can create new experiments to hold new replicates.
