For additional assistance, please contact support@metavilabs.com.
Microscopic Resolution
All FastTrack AI experiments require the user to enter the pixel per µm resolution of the uploaded images (i.e. the number of pixels for each µm of distance in the image). This is important because the FastTrack AI software reports its findings in terms of µm lengths and areas. Without this factor, it would give results in pixels and leave the final conversions to µm to the researcher.
Resolutions lower than 0.4 pix/µm typically do not provide enough visual information for successful analysis. The typical range we see is 0.5 to 2.0 pix/µm.
The Simple Calculation
Please note FastTrack AI uses pix/µm not µm/pix. If you have the resolution in µm/pix, simply invert the result ( 1 divided by the number you have). The (pixel / µm) resolution is calculated by :
The (pixel / µm) resolution is calculated by :
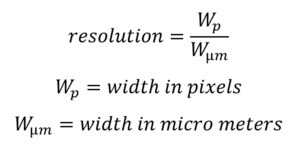
A common mistake is to invert the calculation. If your microscope software provides resolution in µm/pixel, then you can simply apply 1/x to get the resolution in pixels/µm.
Using the embedded Resolution Legend
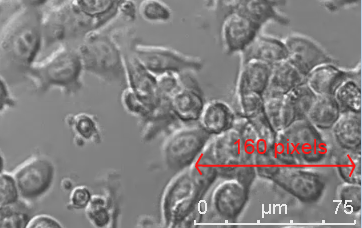
If your microscope embeds a resolution legend then this can be used to determine the pixel / µm resolution very directly. Open the image in a pixel editor such as Paint and measure the number of pixels along the length of the legend. In this example, the resolution is 160 pixels / 75 µm = 2.13 pixel/µm.
Using the Chemotaxis Chamger Walls
If you are using the ibidi chemotaxis chambers, the distance between the chamber walls is known to be 1000 µm. So we can measure the number of pixels from wall to wall using a pixel editor, then calculate the resolution. In this example, it is 1065 pixels / 1000 µm = 1.065 pixels/µm.

Other Resources
This ibidi guide provides more suggestions. Please note the ibidi technical note uses µm/pix whereas FastTrack AI uses pix/µm. So to get the units required by FastTrack AI, simply invert the result ( 1 divided by ibidi-number). ibidi pixel size calculation guide
