
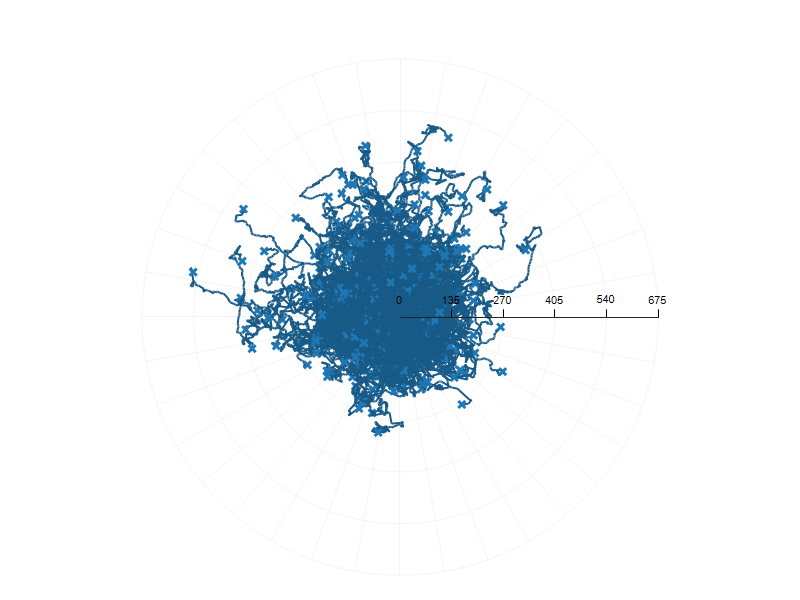
Track thousands of cells - dramatically increase statistical signifigance.
Save many hours of manual tracking time per image sequence.
Increase accuracy and repeatability over human tracking.
Fully compatible with the ibidi Chemotaxis and Migration Tool.
The spontaneous or innate movement of cells as well as the migration induced by certain signal substances (chemokinesis) plays a pivotal role in several physiological, but also in pathological processes. Chemotaxis is a special form of chemokinesis, where cells sense a gradient of a signal substance and move towards it:

MetaVi Labs has developed its chemotaxis analysis platform in close cooperation with ibidi, ensuring that our outputs are fully compatible with the gold-standard methodologies championed by Roman Zantl and the ibidi team. This collaboration means that researchers using MetaVi Labs' software can seamlessly compare, publish, and validate results alongside ibidi's widely adopted µ-Slide Chemotaxis assays and analysis framework.
Both MetaVi and ibidi report FMI as a primary indicator of directional migration. MetaVi computes FMIx and FMIy (aligned with gradient axes), directly paralleling ibidi's FMI‖ and FMI⊥. This allows results to be exchanged and compared without conversion.
MetaVi reports CoMx, CoMy, and magnitude of CoM displacement, fully aligned with ibidi's use of the mean endpoint (M_end) as a measure of population-level bias.
MetaVi calculates the ratio of Euclidean distance to accumulated path length, exactly as ibidi does. This provides a consistent measure of track straightness.
Both systems distinguish cell speed (path-based) from cell velocity (net displacement over time). This prevents confusion between rapid random motility and true chemotaxis.
MetaVi incorporates the Rayleigh test for endpoint uniformity, the same statistical standard recommended by ibidi, ensuring that directional bias is rigorously confirmed.
While remaining 100% compatible with ibidi's framework, MetaVi extends the analysis with additional, complementary metrics:
A diffusion-style metric that provides insights into population motility patterns over time.
User-defined thresholds for track duration and displacement, consistent with ibidi's best practices but adjustable for diverse experimental setups.
Although optimized for ibidi's µ-Slide Chemotaxis chambers, MetaVi software can ingest tracks from virtually any migration assay, making it useful across platforms.
By aligning our analysis platform with ibidi's metrics and methods, MetaVi Labs ensures:
Results can be directly cross-validated with ibidi's Chemotaxis & Migration Tool outputs.
Metrics like FMI, CoM, and Rayleigh test are already recognized in top journals and form the common language of chemotaxis research.
Researchers gain additional insights (e.g. MSD) without sacrificing compatibility.
MetaVi Labs' chemotaxis analysis was built in cooperation with ibidi, making it fully interoperable with ibidi's well-established standards. Researchers can trust that their results will be recognized, reproducible, and directly comparable to the global body of ibidi-based chemotaxis work — with the added power of MetaVi's automation, extended metrics, and device flexibility.
© Copyright 2023